San Jose, June 1st, 2020
InAccel, Inc.
Data scientists, ML engineers and HPC users can now speedup their applications using the power of FPGAs from their browser.
FPGAs are adaptable hardware platforms that can offer great performance, low-latency and reduced OpEx for application like machine learning, video processing, quantitative finance, etc. However, still the easy and efficient deployment from users with no prior knowledge on FPGA was challenging.
InAccel, a pioneer in the domain of application acceleration, has now made possible the easy deployment and utilization of FPGAs from data scientists, ML engineers and HPC users from their browser.
Xilinx™, the world-leader on adaptable platforms, have released as open source the Vitis libraries. Xilinx Vitis™ Library includes an extensive set of open-source, performance-optimized libraries that offer out-of-the-box acceleration with minimal to zero-code changes to your existing applications.
Common Vitis™ accelerated-libraries for Math, Statistics, Linear Algebra, and DSP offer a set of core functionality for a wide range of diverse applications.
Domain-specific Vitis™ accelerated libraries offer out-of-the-box acceleration for workloads like Vision and Image Processing, Quantitative Finance, Database, and Data Analytics, Data Compression and more.
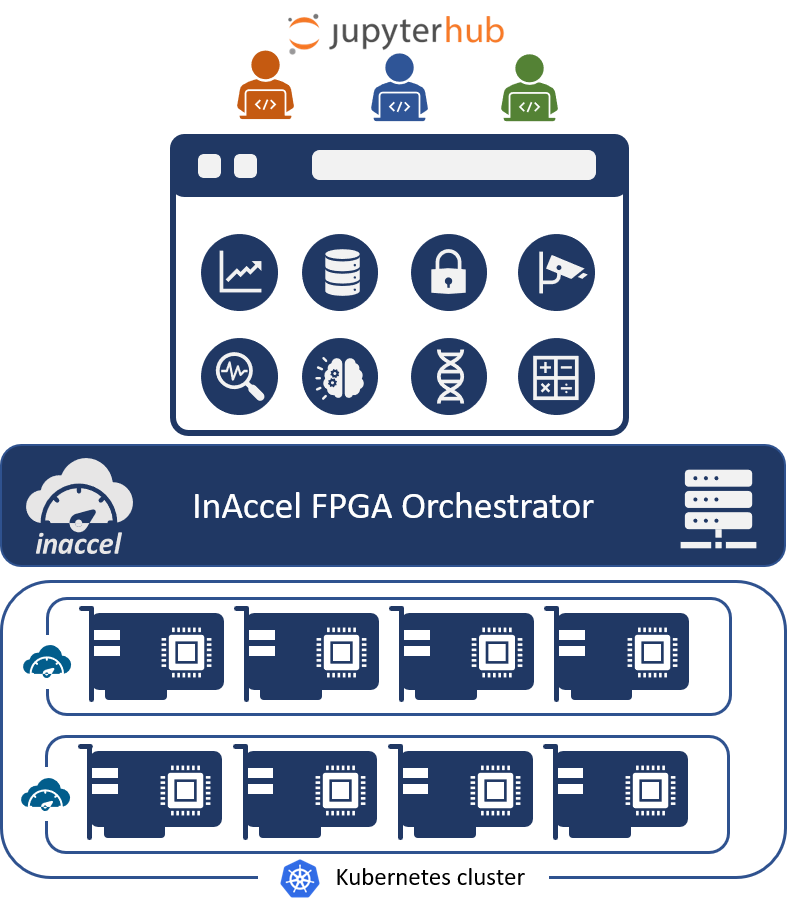
Users now can enjoy the power of the Xilinx™ Vitis™ libraries on the Xilinx™ Alveo™ cards, instantly. InAccel has made available as a demo a framework that allow any user to instantly log in in the platform and start accessing the Xilinx Vitis™ libraries on a cluster of Alveo™ cards. The framework is built on top of Jupyter Hub. InAccel provides an FPGA resource manager that allows the instant deployment, scaling and virtualization of FPGAs making easier than ever the utilization of FPGA clusters for applications like machine learning, data processing, data analytics and many more HPC workloads. User can deploy their application from Python, Jupyter notebooks or even terminals.
Jupyter Notebook is an open-source web application that allows you to create and share documents that contain live code, equations, visualizations and narrative text. Uses include data cleaning and transformation, numerical simulation, statistical modeling, data visualization, machine learning, and much more.
Due to its simplicity it has gained wide popularity among the data scientists and the HPC users that want to create and share applications that integrates the code, the comments and the results of the application.
JupyterHub brings the power of notebooks to groups of users. It gives users access to computational environments and resources without burdening the users with installation and maintenance tasks. Users - including students, researchers, and data scientists - can get their work done in their own workspaces on shared resources which can be managed efficiently by system administrators.
JupyterHub runs in the cloud or on your own hardware, and makes it possible to serve a pre-configured data science environment to any user in the world. It is customizable and scalable, and is suitable for small and large teams, academic courses, and large-scale infrastructure.
Through the JupyterHub integration, users can now enjoy all the benefits that JupyterHub provide such as easy access to computational environment for instant execution of Juputer notebooks. At the same time, users can now enjoy the benefits of FPGAs such lower-latency, lower execution time and much higher performance without any prior-knowledge of FPGAs. InAccel’s framework allows the use of Xilinx’s Vitis Open-Source optimized libraries or 3rd party IP cores (for machine learning, data analytics, genomics, compression, encryption and computer vision applications.)
JupyterHub using InAccel’s FPGA orchestrator can be used either on-prem (e.g. supporting the powerful Xilinx Alveo card) or on the cloud (e.g. AWS, Azure, and Alibaba). That way, users can enjoy the simplicity of the Jupyter notebooks and at the same time experience significant speedups on their applications. InAccel would like to thank the Xilinx CTO organization for their collaboration in support on this initiative.
Users can test for free the available libraries on the InAccel cluster on the following link:
The platform is available for demonstration purposes. Multiple users may access the available cluster with the 2 Alveo cards. If you are interested to deploy your own data center with multiple FPGA cards or run your applications on the cloud, contact us at info@inaccel.com.
