Scikit-learn (also known as sklearn) is a widely used free software machine learning library for the Python programming language. It has been adopted by many companies and universities as it features various classification, regression and clustering algorithms including support vector machines, random forests, gradient boosting, and k-means. SKlearn is designed to interoperate with the Python numerical and scientific libraries NumPy and SciPy.
Scikit-learn is largely written in Python and uses NumPy extensively for high-performance linear algebra and array operations. Furthermore, some core algorithms are written in Cython to improve performance. The vast majority of ready-to-use libraries and the easy deployment has made Scikit-learn a very popular framework across the data science and machine learning community.
However, as it is mostly written in Python and it is designed to be executed in typical processors the performance during the training and the prediction is often limited. Especially for companies that need to process huge amounts of data, the total execution time for the training and the prediction is critical.
Hardware accelerators, like FPGAs, offer high throughput and much better performance compared to typical general-purpose processors (CPUs). However, the programming complexity and the lack of APIs for easy integration with machine learning frameworks was the main barrier for the widespread adoption of FPGAs in machine learning. Currently, FPGAs can be programmed using OpenCL making the development of specialized accelerators much easier. The second constraint was the lack of high-level APIs that allows multiple applications to utilize a cluster of FPGAs.
100x Faster execution time
InAccel was the first company that demonstrated the utilization of FPGAs from high-level frameworks like Spark. It was the first to provide a high-level abstraction of the FPGA resources making easier the deployment from multiple processes or threads. InAccel now offers the required API and the libraries that can be used to speedup up to 100x Scikit-learn applications with zero code changes.
In this case we evaluate the use of InAccel’s Naïve Bayes accelerators integrated with Scikit-learn and deployed in an aws FPGA instance.

Naive Bayes Use case
Naive Bayes classification algorithm is a supervised learning technique, based on applying Bayes’ theorem with the 'naive' assumption that each feature is independent of other features, which can't be true in real life. It is quite popular for its simplicity and is widely used for applications like text classification, sentiment analysis, email filtering, recommender systems and many more. Deeper explanations on how it works are out of scope here, since there are many articles and posts out there discussing the fundamentals of this classifier.
Scikit-learn library, the gold standard for machine learning in Python, offers a set of five different Naive Bayes methods, which differ mainly regarding the data distribution. The end user needs to decide the option that suits better on each specific problem. However, traditional CPUs don't scale so well on bigger data showing a struggle in computational performance. For many applications the performance (mainly latency and throughput) at prediction time of models is crucial. Equally important can be considered the training time, but sometimes it is ignored since it may be done offline in the production.
Gaussian implementation of Naive Bayes underperforms on this stage, when number of features, samples or classes increase and the classification of non-labeled samples using maximum likelihood requires matrix decomposition, a very data-intensive process. So, we show how FPGA acceleration layer can optimize this process.
Speed Test on AWS
The examples were run on AWS EC2 f1 instance equipped with Xilinx VU9P FPGAs and Intel Xeon CPU processors. The datasets were generated with the sklearn.datasets module with different parameters for different synthetic data. More information can be found on this small benchmark.
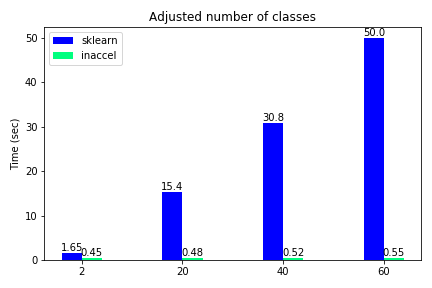
Total execution time using FPGAs for different number of classes
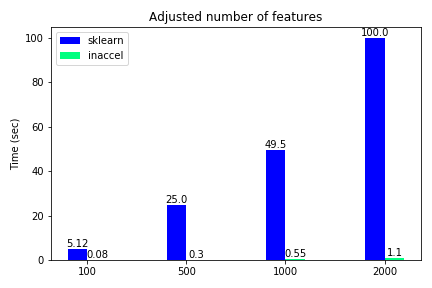
Total execution time using FPGAs for different number of features
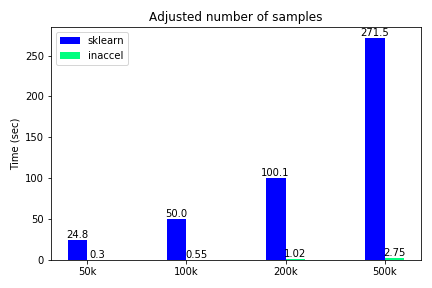
Total execution time using FPGAs for different number of samples
The classification accuracy such as other metrics, are not included in this demonstration, since they were the same on both options and the various use-cases. We notice that only for the binary classification the speedup is relatively 'small'(3.5x) while for all the other cases where the data become more complex, FPGA solution achieves great speedups reaching up to 100x compared to the software-only execution. Moreover, we kept the samples to some thousands and didn’t scale it to millions since the performance improvement is clear.
The most important advantage is that Scikit-learn users can speedup significantly their applications with zero code changes, while also keeping it fully compatible with the rest Scikit-learn. You only need an f1 instance or an FPGA card before following few easy steps to install InAccel API and InAccel Resource Manager.
You can try it to run your own benchmark on Naive Bayes following the instructions in this link
More info of InAccel at: http://inaccel.com
Vangelis Gkiastas
Machine Learning Engineer
InAccel, Inc.
